AI in Medical Imaging
Applying artificial intelligence to predict stroke faster
Overview
Broadly, stroke is when blood flow to a part of the brain is interruped, either via a blood vessel being blocked or bursting. Both of these pathways lead to the death of brain cells, which must be treated extremely quickly. Stroke is one of the leading causes of death annually, with over 7 million fatalities.1 My work here looked at taking patient medical imaging and creating a custom 3D model of their blood vessels, which can allow medical professionals to see where a person may have a stroke. Because my work used patient data, the images will be taken from other sources to protect patient privacy.
The Head CT
Whenever someone is suspected of having a stroke, the first step is typically a head CT scan. For stroke patients, CT scans are mainly perferred over MRI due to the larger presence of CT scanners and the speed at which the imaging can be acquired. For suspected stroke patients, dye is sometimes used that allows radiologists to see blood flow from the imaging
Segmentation
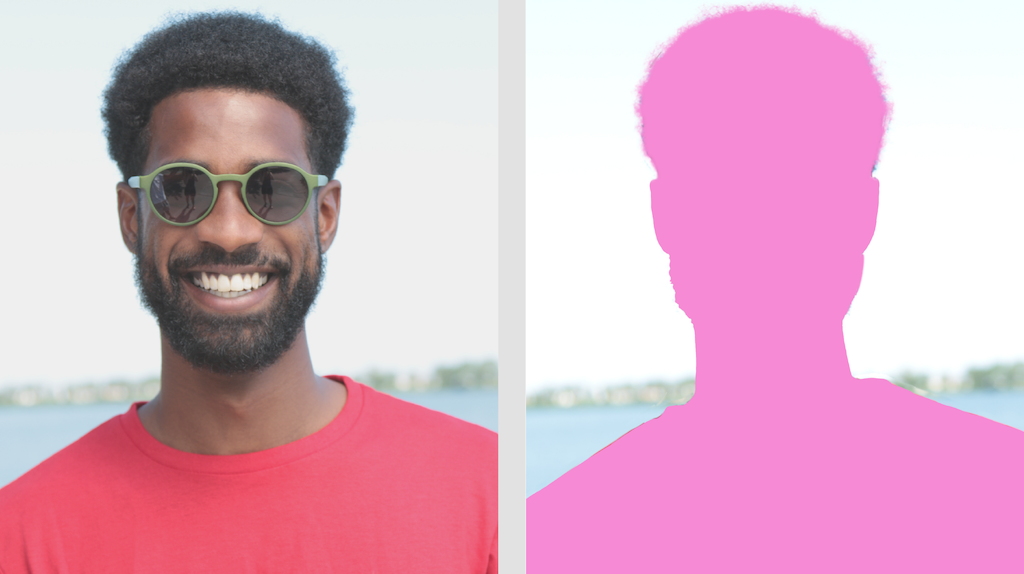
Segmentation in the AI space refers to a model that is able to take an image and divide it into parts. One example would be an image of a person, with the segmented image having identified the person out of the background.
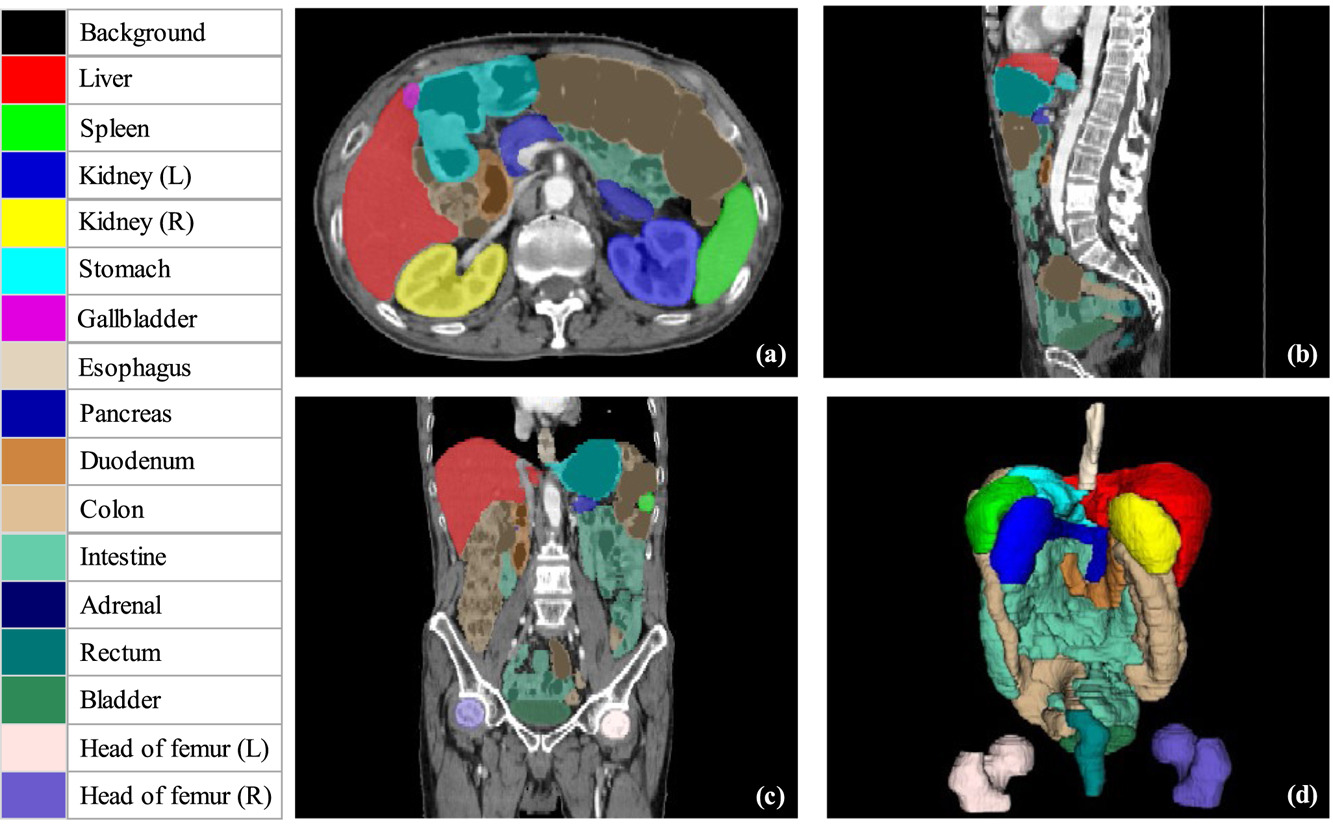
In biomedical imaging, we typically focus on a region of interest depending on what we’re interested in. There are several notable biomedical imaging segmentation tasks, such as tumor segmentation, organ segmentation, or tissue segmentation. An example is given below, with the areas of interest color-coded:
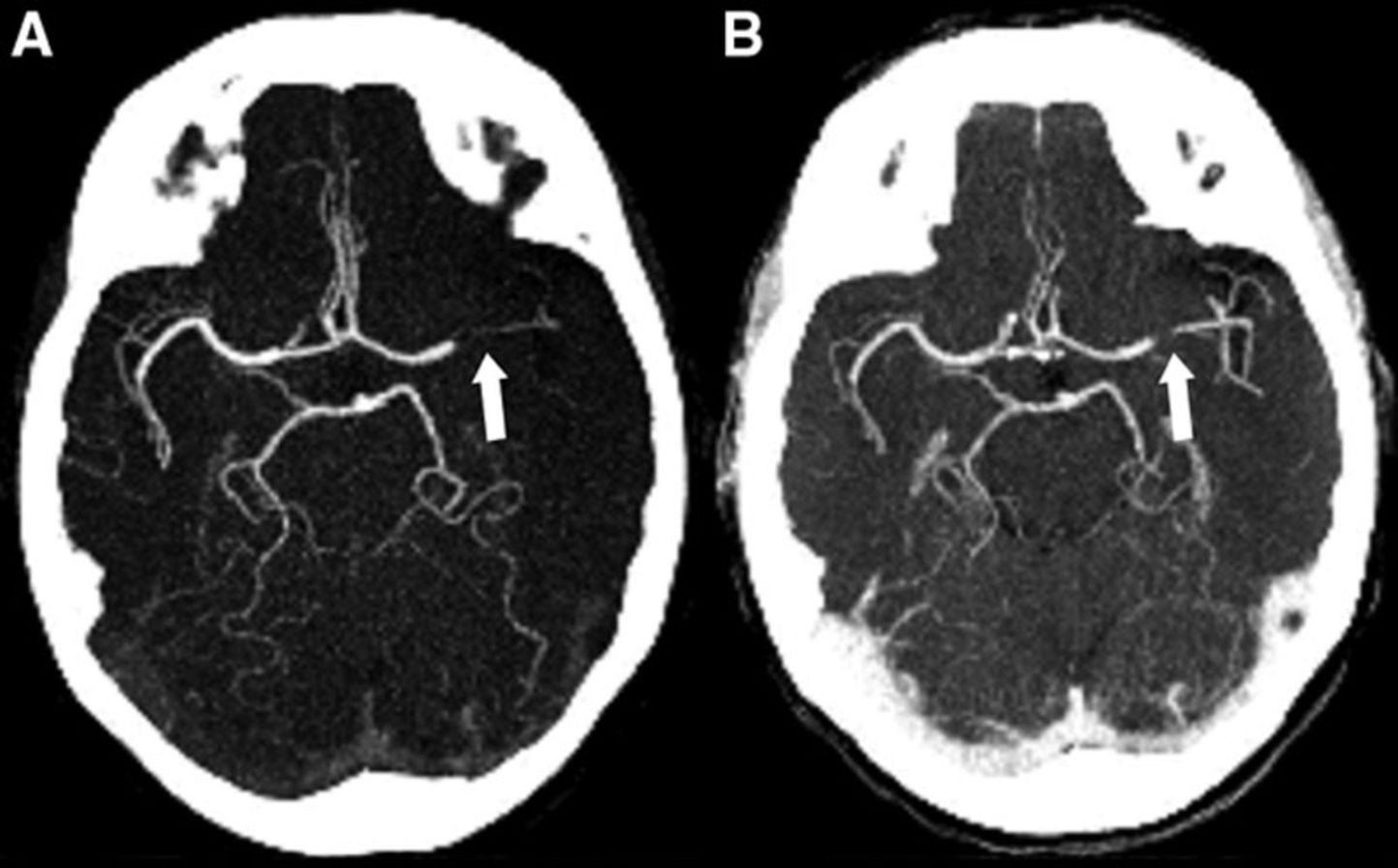
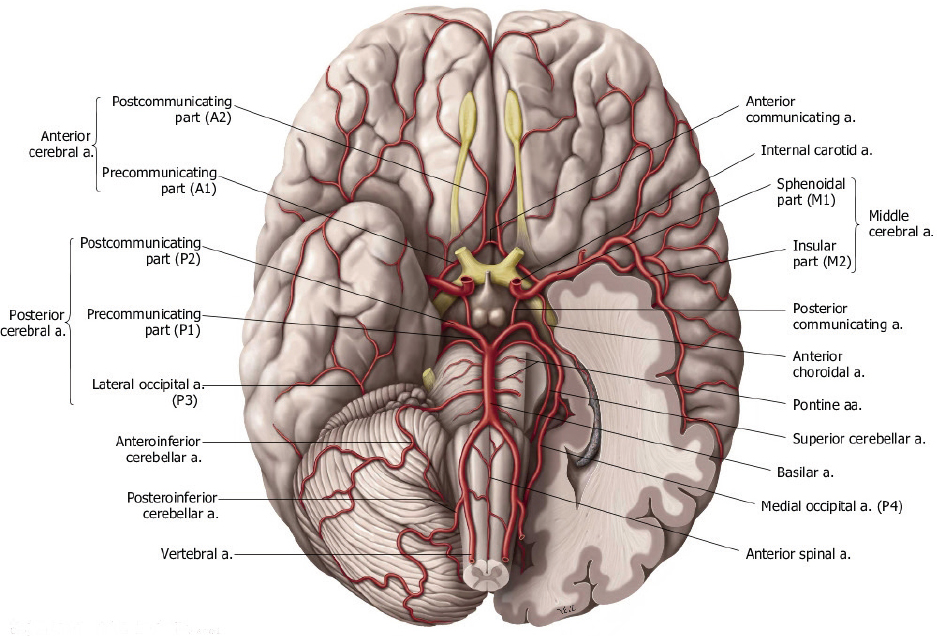
My task was to segment out the blood vessels starting from the top of the heart (specifically the aorta, which is where brain’s blood supply) to the smaller vessels inside of the brain itself (specifically the A2, M2, and P2 segments). These vessels are extremely small (on the order of a few millimeters in diameter).
Biomedical Imaging
Raw biomedical imaging is comprised of three planes (or slices) which allow for the creation of a 3D volume when put together. They are comprised of the axial, coronal, and sagittal planes:
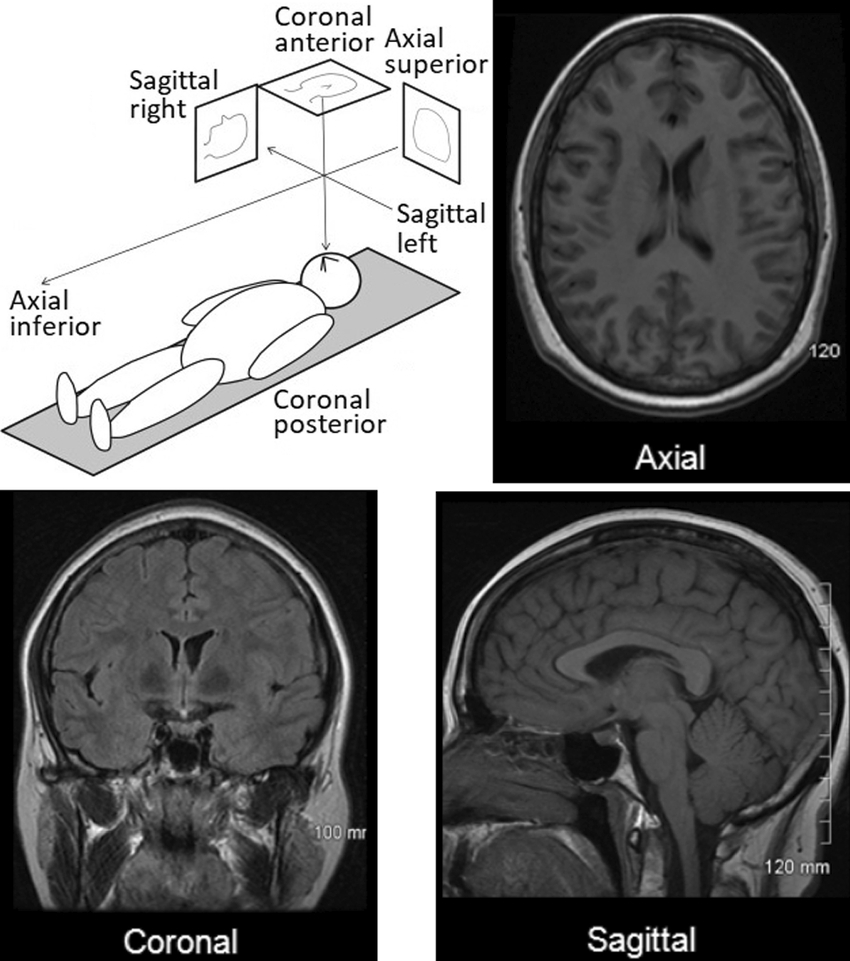
For our segmentation task, the model will be segmenting the blood vessels on each plane, and then stitching all of those predictions together to create our 3D model at the end. This is the same process that the 3D model of the color-coded organs above was made in Figure 1.
Dealing with the Raw Imaging
The next important step is to deal with the imaging formats themselves. Medical imaging formats are typically either DICOM or NIfTI, and contain a lot of important metadata (e.g., the machine used, the spacing between slices, etc.). This metadata means that we can’t simply convert from DICOM/NIfTI to an image format like JPG or PNG, we must keep the imaging in its native format throughout the process. This is exactly where the Medical Open Network for AI (MONAI) comes in.4 It is built from the ground up to keep medical imaging in its native format and make it play nice with AI models.
It also allows us to perform data augmentation natively. From a high level, data augmentation is where we take a small dataset and artificially increase its size by performing random crops, rotations, inject random noise, etc. This causes our dataset size to increase in the hopes that the resulting model will generalize better. Because we are dealing with volume slices, these data augmentations can get pretty messy, but MONAI handles this for us automatically, which is another major plus.
The Model
The most common model used for biomedical image segmentation is U-Net, which is a convolutional neural network (CNN) that was designed specifically for this purpose.5 With the introduction of the transformer architecture, some in the field have begun to turn to so-called Vision Transformers (ViTs) for image segmentation as well.6 7 While I was working on this project, the best-performing segmentation model was the so-called Swin UNETR model, which combined a ViT with a U-Net CNN architecture (creating an ensemble model). ViTs, like text-based transformers such as ChatGPT, are able to keep track of context. This is especially important for our task, as the blood vessels in the brain are going to be present in hundreds of image slices, so it is essential that our network is context aware. @ cite unetr
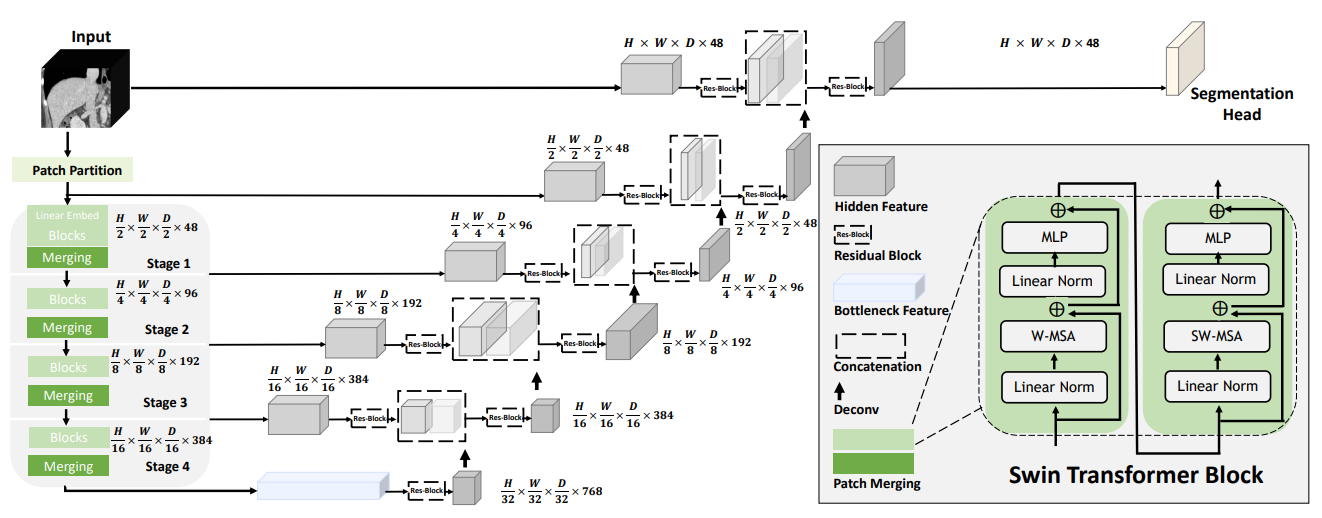
For my work, we elected to use a pre-trained model from the authors and adopt it for our task (a method called transfer learning). This is where the issues began and I learned a ton.
The Problem
According to the authors, the Swin UNETR architecture does extremely well on both the Beyond the Cranial Vault (BTCV) and Medical Segmentation Decathlon (MSD) datasets. The pretrained network we used was trained on about 5000 3D CT images, but none of them were of the brain. Our internal dataset was extremely small and model performance suffered, even with aggressive image augmentation. This led me to a few important lessons (learned the hard way, of course).
The first one is that data is extremely important. Many open-source biomedical segmentation models exist, but these models become fundamentally limited if the data is not there. There are some ways around this, such as using models that have already been trained on large-scale datasets.9
The second one is that of network selection, which I write about a bit here. If someone is applying AI to a task, they should never use only one network. Instead, it is much better practice to pick multiple networks, apply them to your task, and pick the one that ends up performing the best.
References
Citation
@online{gregory2025,
author = {Gregory, Josh},
title = {AI in {Medical} {Imaging}},
date = {2025-09-30},
url = {https://joshgregory42.github.io/projects/imaging/},
langid = {en}
}