Modeling Blood Clots Dynamically
Recreating blood clot fluid dynamics in silico from intravital microscopy imaging.
A lot of this project can be found in the paper published by the lab and other collaborators.1 All data presented here is either from that paper or public conferences. See the bottom of this page for the citation.
Blood Clots are Dynamic
Many people (myself included at the beginning of this project) view blood clots as static and solid objects. In my head, I thought of blood clot blockages (the technical term here is thrombus or thrombi for plural) as these little rocks that sat in your blood vessels. This is very much not the case. After vessel injury, there are many dynamic processes that begin to take place. The clot itself continuously deforms and grows over time, interacting with the flow environment around it. Furthermore, the structure of a blood clot itself is very heterogeneous and exhibits a core-shell structure.2 3 The core of a clot consists mainly of dense, highly activated platelets and fibrin, surrounded by a shell of looser, less activated platelets and fibrin. Here, “activated” and “less activated” indicate the tendency of the platelets to adhere to themselves. This in turn leads to a higher chance of a thrombus forming.
Study Aim
In this work, I worked with a PhD student to combine in vivo and in silico methods to simulate heterogeneous blood clots transiently with computational fluid dynamics (CFD). We worked with experimentalists (specifically Maurizio Tomaiuolo and Timothy J. Stalker from Thomas Jefferson University) to obtain the imaging, which is described next.
Study Arms
Two kinds of mice models were used for this work: standard wild type (WT) mice that clot normally, and mice with a clotting factor removed, specifically a substitution of phenylalanine for two tyrosine residues in the \(\alpha_{IIb} \beta_3\) glycoprotein, referred to as diYF knockout mice. The rationale behind these two arms was to show that the resulting in silico methods are able to distinguish the resulting forces on the blood clot, with the hypothesis being the clots in the WT mice will have larger forces present than the diYF due to the lack of a clotting factor present in the diYF mice.
Mice Imaging
The time series mice imaging was taken via intravital microscopy of laser injury experiments in mouse cremaster muscle arteriole. The laser was used to puncture the vessel lumen in live mice, which began the clotting process.
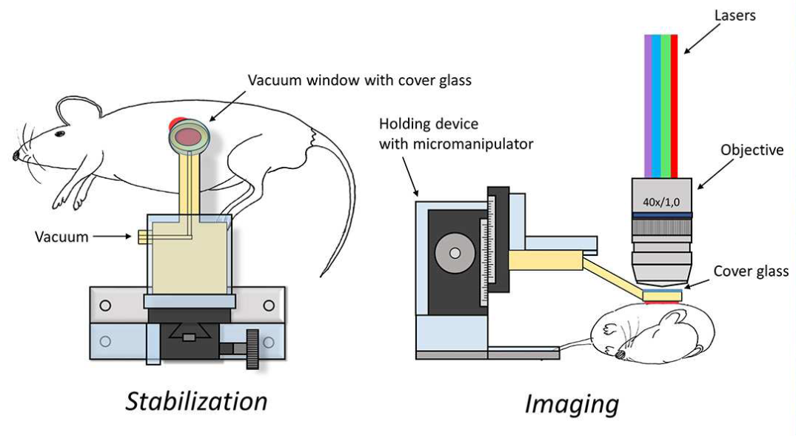
During this process, imaging captured signal intensity of three flourescent tracers: caged flourescent albumin (cAlb), P-Selectin, and CD41. Briefly, cAlb indicates blood plasma and its leakage from the clot; P-Selectin indicates the high density portion of the clot near the injury site, mainly consisting of activated platelets; CD41 is associated with platelets that form the solid part of the blood clot. Each experiment consists of 300 images taken at intervals of 0.55 seconds.
In Silico Workflow
The resulting in silico methodology developed here is called Integrated In Silico Modeling, or IVISim.
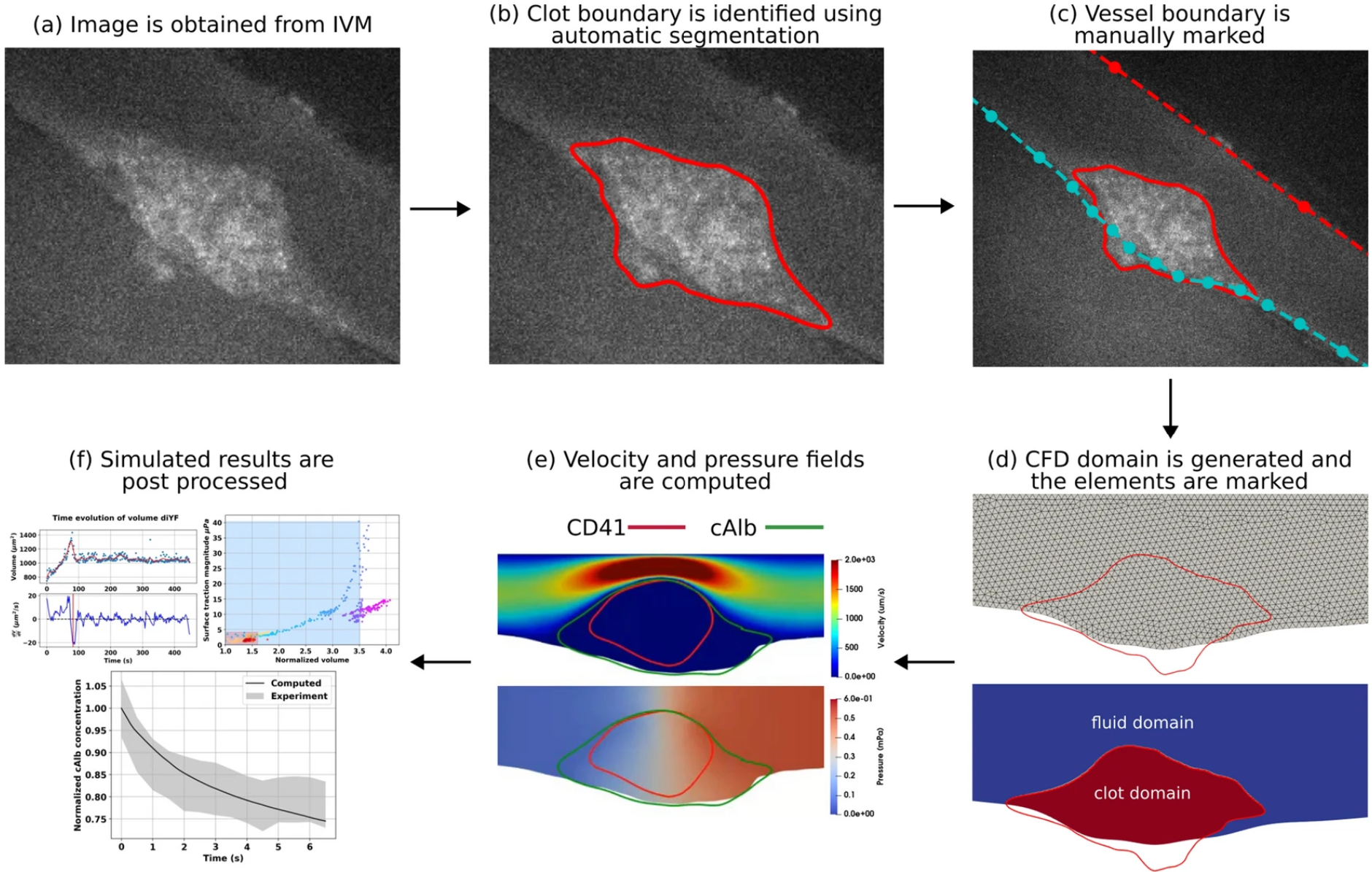
From a high level, the IVISim workflow involves the following steps (following #fig-ivisim-workflow):
- The intravital microscopy imaging is obtained from the experimentalists, with each image pre-processed to remove noise.
- Clot boundary is segmented across all frames
- The top and bottom of the vessel is marked manually from a single frame, then applied to all frames in the stack.
- The clot domain and fluid domain is then marked.
- CFD simulations are then run to compute the velocity and pressure fields for each frame.
- These results are then post-processed to obtain measurements such as clot force and shape.
The below video shows a completed simulation after being passed through the IVISim workflow. The yellow region corresponds to the CD41 (clot core) channel, and the red region corresponds to the cAlb (blood plasma) channel. The blood flow direction is from left to right.
From this video, we can see how dynamic a blood clot is over time. We can see how it grows, taking up a larger percentage of the vessel, then eventually shrinks down, reaching a steady-state shape.
The Paper
For more details, please refer to the publication by Teeraratkul and colleagues at the bottom of this page.1
References
Citation
@online{gregory2025,
author = {Gregory, Josh},
title = {Modeling {Blood} {Clots} {Dynamically}},
date = {2025-10-07},
url = {https://joshgregory42.github.io/projects/clot-modeling/},
langid = {en}
}